GenePattern users can leverage Globus transfers for analysis of large biomedical datasets
July 07, 2022 | Brigitte Raumann
With the increasing prevalence of new ‘omics technologies such as single-cell RNA-seq and spatial transcriptomics, as well as multi-omic analysis methods that integrate datasets from different ‘omics domains, bioinformatics tools are required to operate on ever larger datasets, which can range in the tens to hundreds of gigabytes or greater. One such bioinformatics tool is GenePattern, developed in the Mesirov Lab at the School of Medicine at UCSD. GenePattern provides a platform for reproducible genomic analysis for users at all levels of computational sophistication. The application presents scientists with hundreds of analyses, including general machine learning approaches, ‘omic platform-specific methods, cancer-focused analyses, and essential utilities. Analyses can be chained into workflows that are shareable, publishable, and reproducible.
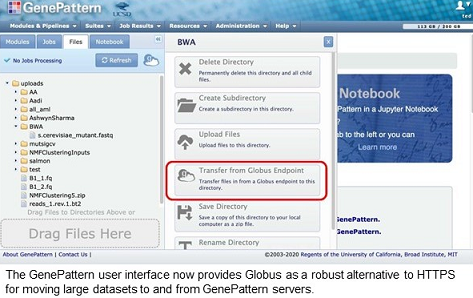
The GenePattern server is a well-established bioinformatics analysis resource, with over 85,000 registered users running up to 10,000 analyses per month, most of which require the upload of datasets and subsequent download of result files. Researchers can upload data to GenePattern servers via the standard protocol for web servers, HTTPS. However, as the size of datasets grows, some users are encountering the limitations of HTTPS-based transfers, such as buffering size limits in browsers and web servers and transfer failures due to browser timeouts, brief network outages, browser session interruptions, and endpoint disconnection.
With funding from the Informatics Technology for Cancer Research program of the National Cancer Institute, the GenePattern and Globus teams have collaborated to integrate Globus data transfer capabilities into the GenePattern user interface in order to provide researchers with a high performance alternative to HTTPS data transfer. Now GenePattern users have a robust, secure file transfer option for moving large datasets to and from public GenePattern servers hosted on the Amazon cloud or the Expanse high performance compute cluster at the San Diego Supercomputer Center through the Extreme Science and Engineering Discovery Environment (XSEDE) program.
The collaboration's next step is to incorporate Globus file transfer capabilities into the GenePattern Notebook JupyterLab interface, allowing researchers to perform GenePattern analyses of large datasets in the JupyterLab environment.
Learn more about the Globus platform
Related Content
Streamlining Reproducible Genomics Analysis with GenePattern and the Globus Platform
UCSD School of Medicine
Researchers needed an easy to use platform for reproducible bioinformatics research so the GenePattern Gateway for Genomic Analysis was developed....